In this paper, we generate 3D visual explanations to analyze the 3D brain tumor segmentation model by extending a post-hoc interpretability technique.
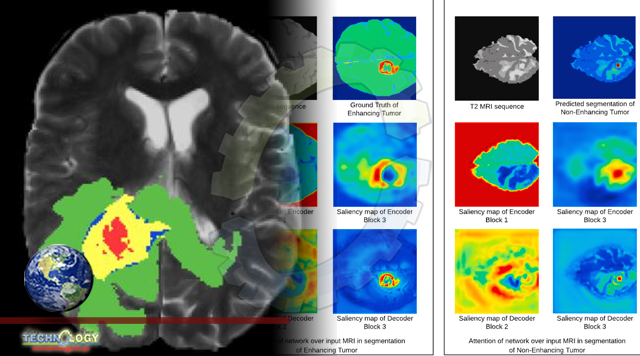
Medical image segmentation is a complex yet one of the most essential tasks for diagnostic procedures such as brain tumor detection. Several 3D Convolutional Neural Network (CNN) architectures have achieved remarkable results in brain tumor segmentation.
However, due to the black-box nature of CNNs, the integration of such models to make decisions about diagnosis and treatment is high-risk in the domain of healthcare.
It is difficult to explain the rationale behind the model’s predictions due to the lack of interpretability. Hence, the successful deployment of deep learning models in the medical domain requires accurate as well as transparent predictions.
In this paper, we generate 3D visual explanations to analyze the 3D brain tumor segmentation model by extending a post-hoc interpretability technique.
We explore the advantages of a gradient-free interpretability approach over gradient-based approaches. Moreover, we interpret the behavior of the segmentation model with respect to the input Magnetic Resonance Imaging (MRI) images and investigate the prediction strategy of the model.
We also evaluate the interpretability methodology quantitatively for medical image segmentation tasks. To deduce that our visual explanations do not represent false information, we validate the extended methodology quantitatively.
We learn that the information captured by the model is coherent with the domain knowledge of human experts, making it more trustworthy. We use the BraTS-2018 dataset to train the 3D brain tumor network and perform interpretability experiments to generate visual explanations.
Originally published at Physicians weekly